
Full text loading...
A converting phage for ampicillin resistance - phage 5006Mpa - was used to transduce Proteus mirabilis strain pm5006 to ampicillin resistance. The infecting phage carried TnI as the source of its phenotype. The recipient had the conjugative plasmid P-lac as well as a non-self-transmissible plasmid as residents. The latter was a recombinant between a morganocinogenic plasmid Mor174 and R plasmid R772 which coded for kanamycin resistance. This recombinant plasmid had possibly undergone transductional shortening as a result of previous uptake by transducing phage 5006M. From these transductants, three transducing systems for transfer of morganocinogeny were obtained. The first consisted of the complete transduction of a plasmid expressing markers of Mor174 and ampicillin resistance at frequencies of about 1 × 10−5 per phage particle adsorbed. This frequency could have equalled that of the adsorption of transducing phage to recipient cells. The transduced plasmid was formed by translocation of Tn1 to the Mor174R772 complex with inactivation of the kanamycin resistance marker of the latter. The transducing phage was named phage 5006Mmora. The second - a high frequency transducing system for morganocinogeny, kanamycin and ampicillin resistances - was the result of markers of the Mor174R772 complex inserting contiguously to Tn1 in the 5006Mpa-pm5006 cryptic prophage sequence. The insertion converted the terminally redundant, circularly permuted phages into deficient phages lacking some (not always the same) genes and being unable to circularize (and hence lysogenize) due to lack of terminal repetition. The recombination of two defective genomes resulted in doublets which could reduce to the prophage state. This mutual aid explained the phage multiplicity dependence phenomenon encountered with this system. The phage was named phage 5006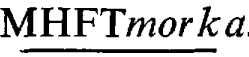 The third system of transduction resulted from deletion of non-essential genes from the oversized genomes just described. This restored terminal redundancy and consequently allowed individual genomes to circularize and thus transduce the three markers. This phage was named phage 5006Mdpmorka. These phages also transduced their markers to P. mirabilis
pmoxk, a strain to which only slight and variable adsorption of the phage could be demonstrated. Properties of the systems are described.
The third system of transduction resulted from deletion of non-essential genes from the oversized genomes just described. This restored terminal redundancy and consequently allowed individual genomes to circularize and thus transduce the three markers. This phage was named phage 5006Mdpmorka. These phages also transduced their markers to P. mirabilis
pmoxk, a strain to which only slight and variable adsorption of the phage could be demonstrated. Properties of the systems are described.

Article metrics loading...

Full text loading...
References


Data & Media loading...
